Scientists develop model for more efficient simulations of protein interactions linked to cancer
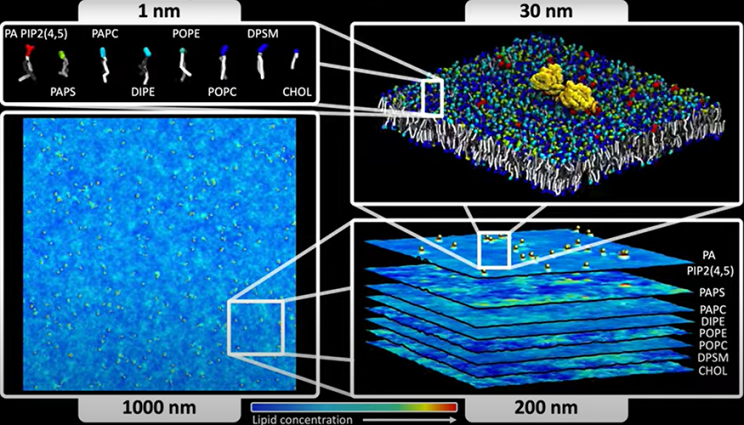
LLNL scientists have developed a theoretical model for more efficient molecular-level simulations of cell membranes and their lipid-protein interactions, part of a multi-institutional effort to better understand the behavior of cancer-causing membrane proteins. Developed under an ongoing collaboration by the Department of Energy and the National Cancer Institute (NCI) aimed at modeling cell membrane interactions with RAS—a protein whose mutations are tied to about 30% of human cancers—the new model addresses a major problem in simulating RAS behavior, where conventional methods come up well short of reaching the time- and length-scales needed to observe biological processes of RAS-related cancers. The work appears in the latest issue of the journal Physical Review Research. The new model, based on Dynamic Density Functional Theory (DDFT), enables simulations that can access micron-level length-scales and timescales on the order of seconds, while maintaining resolution close to the current gold standard of molecular dynamics (MD) models. For perspective, these scales are hundreds of times larger in space and many thousands of times longer in time than those accessible with MD. Read more at LLNL News, which includes a brief video of the model's process.