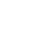The DSI’s Open Data Initiative (ODI) enables us to share LLNL’s rich, challenging, and unique datasets with the larger data science community. Our goal is for these datasets to help support curriculum development, raise awareness around LLNL’s data science efforts, foster new collaborations, and be leveraged across other learning opportunities.
As we develop this catalog over time, the data will represent a wide variety of key LLNL mission areas and may include subsets of some of the world’s largest datasets. We plan to provide data ranging in complexity from dense, featureful, labeled datasets with well understood solutions to those that are sparse, noisy, and largely unexplored. These datasets can also be used to test novel hardware solutions for scalable machine learning platforms.
Download (LLNL-MI-84834) | License
In fluid mechanics problems, computational fluid dynamics (CFD) uses data structures and numerical analysis to investigate the flow of liquids and gases. Researchers in LLNL’s Atmospheric, Earth, and Energy Division use CFD models to simulate atmospheric transport and dispersion. For instance, simulations of wind-driven dynamics can be used to train machine learning models that, in turn, can predict spatial patterns with high accuracy.
This dataset includes 16,000 CFD simulations—15,000 training cases and 1,000 test cases—post-processed for machine learning training. The physics problem is a 2D spatial pattern formed from a pollutant that has been released into the atmosphere and dispersed for up to an hour while undergoing deposition to the surface. The pollutant’s release location is assumed to occur anywhere in a 2D domain of 5000 × 5000 meters. The release is initialized from a small bubble centered 5 meters above the surface, has a radius of 5 meters, and has internal momentum that causes it to expand within the initial minute of simulation time. All the realizations use unit mass releases, and the resulting deposition patterns can be scaled proportionately for other mass amounts. The pollutant is blown in a direction controlled by the large-scale atmospheric inflow winds expressed with variable wind speeds. The goal is to predict a deposition image given its associated release location and wind velocity. The research was funded by the National Nuclear Security Administration, Defense Nuclear Nonproliferation Research and Development (NNSA DNN R&D).
References: (1) Predicting wind-driven spatial deposition through simulated color images using deep autoencoders; (2) Large eddy simulations of turbulent and buoyant flows in urban and complex terrain areas using the Aeolus model
View all datasets in the UCSD LLNL collection.
Code repository (LLNL-CODE-764041) | Download (LLNL-MI-835833) | License
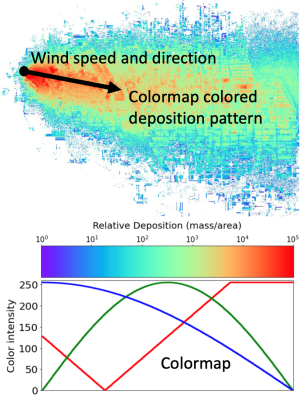
Building off LLNL's Cardioid code, which simulates the electrophysiology of the human heart, a research team has conducted a computational study to generate a dataset of cardiac simulations at high spatiotemporal resolutions. The dataset, which is publicly available for further cardiac machine learning research, was built using real cardiac bi-ventricular geometries and clinically inspired endocardial activation patterns under different physiological and pathophysiological conditions.
The dataset consists of pairs of computationally simulated intracardiac transmembrane voltage recordings and electrocardiogram (ECG) signals. In total, 16,140 organ-level simulations were performed on LLNL's Lassen supercomputer, concurrently utilizing 4 GPUs and 40 CPU cores. Each simulation produced pairs of 500ms-by-12 ECG signals and 500ms-by-75 transmembrane voltage signals. The data was conveniently preprocessed and saved as NumPy arrays.
This project was funded by the Laboratory Directed Research and Development program (18-LW-078, principal investigator: Robert Blake), and the paper "Intracardiac Electrical Imaging Using the 12-Lead ECG: A Machine Learning Approach Using Synthetic Data" was accepted to the 2022 Computing in Cardiology international scientific conference. Co-authors are LLNL's Mikel Landajuela, Rushil Anirudh, and Robert Blake along with Joe Loscazo from Harvard Medical School.
View all datasets in the UCSD LLNL collection.
Download (LLNL-CONF-830547) | License
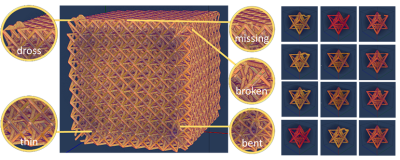
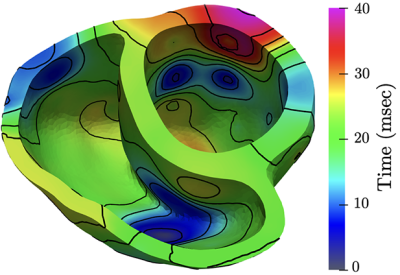
Computed Tomography (CT) is a common imaging modality used in industrial, healthcare, and security settings. Researchers at LLNL use it for non-destructive evaluation in a wide range of applications, including materials science and manufacturing processes. For example, CT imaging can highlight defects in additively manufactured structures, which aids in fine-tuning subsequent iterations of development.
These seven datasets are simulations, containing input and output models of x-ray CT simulations showing additively manufactured lattice structures with common defects. Each dataset contains an input file—computer-aided design triangle mesh model in *.obj format—and an output file—a 3D numpy array, in *.npy format, where each value corresponds to an x-ray CT density value. The input files were created using Blender, and five defects were manually inserted: bent strut, broken strut, missing strut, thin strut, and dross defect. The output files are simulations created using Livermore Tomography Tools (LTT), a software package that includes all aspects of CT modeling, simulation, reconstruction, and analysis algorithms based on the latest research in the field. The data was collected during Joseph Tringe’s Laboratory Directed Research and Development Strategic Initiative project (20-SI-001).
View all datasets in the UCSD LLNL collection.
Download (LLNL-MI-784762) | License
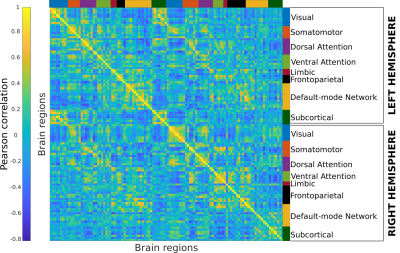
The Human Connectome Project–Young Adult (HCP-YA) dataset includes multiple neuroimaging modalities from 1,200 healthy young adults. These modalities include functional magnetic resonance imaging (fMRI), which measures the blood oxygenation fluctuations that occur with brain activity.
The fMRI data were recorded in multiple sessions per subject: during rest and a set of tasks, designed to evoke specific brain activity. Each fMRI run is a sequence of 3D volumes, and processing these large collections of data is computationally expensive. LLNL researchers have processed these time-series and generated the hierarchically parcellated connectomes using high-performance computing resources. Using these data, LLNL and Purdue University researchers have assessed the “brain fingerprint” gradients in young adults by developing an extension of the differential identifiability framework. For more information, see the paper Functional Connectome Fingerprint Gradients in Young Adults.
This dataset contains timeseries parcellated using the Schaefer parcellations at resolutions ranging from 100 to 900 brain regions. The fMRI data contain resting state and task data for both left-to-right and right-to-left acquisition polarity. This release also contains outputs with and without Global Signal Regression (GSR). In addition to the timeseries data, correlation-based functional connectome (FC) matrices are also included for both GSR and non-GSR.
View all datasets in the UCSD LLNL collection.
Code repository (LLNL-CODE-825407)

ZTF is a wide-field astronomical imaging survey seeking to answer many of the universe's mysteries, by frequently imaging billions of objects in our night sky. Understanding the nature of dark matter and dark energy relies on accurately mapping the universe around us, making distinguishing stars from galaxies a crucial task for astronomers. Mislabeling stars and galaxies poses a considerable problem for cosmological models, and current methods for separating these populations consumes valuable time and resources, driving astronomers to search for new methods of deciphering objects in a quick and effective manner.
Scientists at LLNL have developed a computationally efficient Gaussian process hyperparameter estimation method, MuyGPS, that can be applied to image classification tasks. This repo demonstrates the use of MuyGPyS, a python package that implements the MuyGPS method, and contains various descriptive notebooks covering everything from curating a dataset of real stars and galaxies, to obtaining a classification accuracy. For more information:
Dynamic computed tomography (DCT) refers to reconstruction of moving or non-rigid objects over time while x-ray projections are acquired over a range of angles. The measured x-ray sinogram data represents a time-varying sequence of dynamic scenes, where a small angular range of the sinogram will correspond to a static or quasi-static scene, depending on the amount of motion or deformation as well as the system setup. The reconstruction of DCT is widely applicable to the study of object deformation and dynamics in a number of industrial and clinical applications (e.g., heart CT). In material science and additive manufacturing applications, the DCT capabilities aid in the study of damage evolution due to dynamic thermal loads and mechanical stresses over time which provides crucial information about their overall performance and safety.
We provide two dynamic CT datasets (D4DCT-DFM, D4DCT-AFN) where the sinogram data represent a time-varying object deformation to demonstrate damage evolution due to several mechanical stresses (compression). The provided datasets enable training and evaluation of the data driven machine learning methods for DCT reconstruction. To build the datasets, we used Material Point Method (MPM)-based methods to simulate deformation of objects under mechanical loading, and then simulated CT sinogram data using Livermore Tomography Tools (LTT).
View all datasets in the UCSD LLNL collection.
Labeled dataset | Raw dataset (2263MB each)
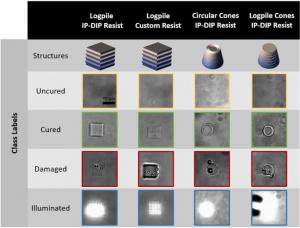
Two-photon lithography (TPL) is a widely used 3D nanoprinting technique that uses laser light to create objects. Challenges to large-scale adoption of this additive manufacturing method include identifying light dosage parameters and monitoring during fabrication. A research team from LLNL, Iowa State University, and Georgia Tech is applying machine learning models to tackle these challenges—i.e., accelerate the process of identifying optimal light dosage parameters and automate the detection of part quality. Funded by LLNL’s Laboratory Directed Research and Development Program, the project team has curated a video dataset of TPL processes for parameters such as light dosages, photo-curable resins, and structures. Both raw and labeled versions of the datasets are available via the links above.
Learn more:
- “Machine learning model may perfect 3D nanoprinting,” LLNL News, July 29, 2020.
- X.Y. Lee, S.K. Saha, S. Sarkar, B. Giera. “Automated detection of part quality during two-photon lithography via deep learning,” Additive Manufacturing 36, December 2020. doi.org/10.1016/j.addma.2020.101444
- X.Y. Lee, S.K. Saha, S. Sarkar, B. Giera. “Two Photon lithography additive manufacturing: Video dataset of parameter sweep of light dosages, photo-curable resins, and structures,” Data in Brief 32, October 2020. doi.org/10.1016/j.dib.2020.106119
Data portal | Project information
A public data portal provides a wealth of data LLNL scientists have gathered from their ongoing COVID-19 molecular design projects, particularly the computer-based “virtual” screening of small molecules and designed antibodies for interactions with the SARS-CoV-2 virus for drug design purposes. The data is queryable by criteria such as chemical structure and binding probability scores, so outside researchers can easily locate relevant data for their own work. The portal is regularly updated and will provide the results of experiments performed at the Laboratory on the effectiveness of small molecules and antibodies against SARS-CoV-2.
CORD-19 dataset | Visualization dashboard

An LLNL research team has extracted drug compounds and therapeutic agents mentioned in the COVID-19 Open Research Dataset (CORD-19 as of June 22, 2020) using natural language processing. The extracted information is provided in an interactive data visualization dashboard (via Tableau) for researchers to explore which drug compounds have been used to treat and/or study coronavirus/COVID-19. With this interactive tool, researchers studying treatments for COVID-19 can quickly visualize the drug compounds that have been studied, the combination of drugs that have been explored, and the classes of drugs that are being mentioned in scientific literature.
Learn more:
- COVID-19 Open Research Dataset informational page on main LLNL website
- Instructions for using the Tableau visualization dashboard
- ChEMBL database for individual compounds
- Anatomical therapeutic codes used by the World Health Organization
Code repository (LLNL-CODE-808183) | Download (LLNL-MI-813373)
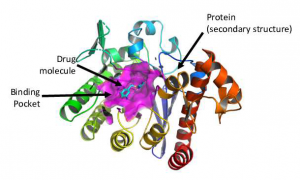
This dataset contains protein-ligand complexes in a 3D representation for anti-viral drug screening against SARS-CoV-2. This is a part of the LLNL COVID-19 database but is specifically designed to facilitate machine learning and other data science tasks with regard to both efficacy (protein-ligand binding affinity) and safety. This complex dataset is called ml-hdf, comprised of ligands and four potential binding pockets of the SARS-CoV-2 protein targets in a 3D atomic representation. The ligands in this dataset includes U.S. Federal Drug Administration (FDA) approved drugs and "Other-world-approved" drugs that have been approved for use by the European Union, Canada, and Japan. The compounds were docked against two binding pockets from the Spike protein (spike, spike1) and two conformations of the main protease (protease, protease2).
View all datasets in the UCSD LLNL collection.
Code repository | Download (LLNL-MI-811381)

The dataset comprises trace files from high-performance computing (HPC) simulations. The trace files contain records of every I/O operation executed by a simulation application run, including I/O operations from HDF5, MPI-IO, and POSIX and all of the parameters supplied to those operations (e.g., file name, offset, and flags). The traces are generated by executing a simulation application that is linked with the Recorder tracing tool. The Recorder tracing tool intercepts the I/O calls made by the application, records the I/O trace record, and then calls the intended I/O call so that the operation executes.
View all datasets in the UCSD LLNL collection.
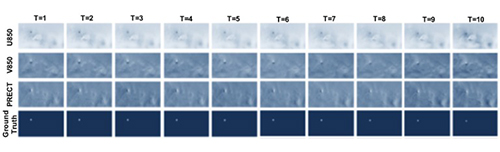
This dataset includes 20-year-long records from 1996 to 2015 of the Community Atmospheric Model v5 (CAM5) dataset. It contains snapshots of the global atmospheric states for every 3 hours (1 timestep = 3 hours). Each snapshot contains multiple physical variables among which we use the six most important climate variables to define hurricane from scientific literature:
- PSL (Sea level pressure)
- U850 (Zonal wind)
- V850 (Meridional wind)
- PRECT (Precipitation)
- TS (Surface temperature)
- QREFHT (Reference high humidity)
View all datasets in the UCSD LLNL collection.
Code repository | Download (LLNL-MI-699521)

The Cars Overhead With Context (COWC) dataset is a large set of annotated cars from overhead. It is useful for training a device such as a deep neural network to learn to detect and/or count cars. More information is available via the researchers’ paper and poster.
The data includes wide area imagery with annotations as well as precompiled image sets for training/validation of classification and counting. The dataset and research to create this data was done by members of the Computer Vision group within LLNL’s Computation Engineering Division under grant from NA-22 in the Global Security Directorate.
The dataset has the following attributes:
- Data from overhead at 15 cm per pixel resolution at ground.
- Data from 6 distinct locations: Toronto, Canada; Selwyn, New Zealand; Potsdam and Vaihingen, Germany; Columbus, Ohio, USA; and Utah, USA.
- 32,716 unique annotated cars. 58,247 unique negative examples.
- Intentional selection of hard negative examples.
- Established baseline for detection and counting tasks.
- Extra testing scenes for use after validation.
View all datasets in the UCSD LLNL collection.
Code repository (LLNL-CODE-772361) | Download

The JAG model has been designed to give a rapid description of the observables from inertial confinement fusion (ICF) experiments, which are all generated very late in the implosion. This code contains pre-trained ML models, architectures and implementations for building surrogate models in scientific ML. The provided dataset is intended for testing/training the models. It is a tarball inside 'data/', which contains .npy files for images, scalars, and the corresponding input parameters. This 10K dataset represents a larger 100M dataset and is beginning to push the 1B threshold with more to come.
View all datasets in the UCSD LLNL collection.