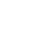Sept. 22, 2021
4D Computed Tomography Reconstructions
Computed tomography (CT) is a type of x-ray imaging technology with a range of applications for clinical diagnosis, non-destructive evaluation in industry, baggage inspection, and cargo screening. CT scanners capture a sequence of angles around an object. Reconstruction algorithms then estimate the scene from these measured images. 2D and 3D CT imaging of static objects are well-studied problems with theoretical and practical algorithms. However, reconstruction of scene changes and measurements over time, known as dynamic 4D CT, can yield spatiotemporal ambiguities. (Image at left: 4D CT of material deformation at beginning and end stages [left] and human thoracic cavity at three different breathing phases [right]. From top to bottom: ground truth, scene reconstruction with the research team's novel 4D CT method, and existing state-of-the-art method.)
Together with Arizona State University colleagues, LLNL researchers have designed a reconstruction pipeline using implicit neural representations (INR, i.e., use of deep learning techniques for optimization) with a novel parametric motion field warping to perform 4D-CT reconstruction of rapidly changing scenes of moving or deformable objects with limited view samplings. The team's model reconstructs a dynamic scene as a static scene coupled with a parametric motion field to estimate an evolving 3D object over time; then the reconstruction is synthesized into image measurements using mathematical transforms that simulate CT scanners. This approach minimizes the discrepancy between the synthesized and observed images, enabling optimization of both the INR weights and motion parameters to obtain accurate dynamic reconstructions without training data. Benchmarked on several datasets including thoracic images (shown above), the algorithm has proven to accurately reconstruct the motion and geometry of the imaged objects through time.
The team’s paper, titled “Dynamic CT Reconstruction from Limited Views with Implicit Neural Representations and Parametric Motion Fields,” has been accepted to the 2021 International Conference on Computer Vision (ICCV), October 11–17. For more information, contact hkim [at] llnl.gov (Hyojin Kim).

LVOC Expansion Supports Predictive Biology Research
Two new buildings at the Livermore Valley Open Campus (LVOC) opened in August, providing office and meeting space for LLNL researchers in predictive biology, materials, and manufacturing, as well as the Innovation & Partnerships Office and the High Performance Computing Innovation Center (HPCIC). Sitting on 110 acres just outside the Laboratory gates, LVOC is a joint initiative of the NNSA, LLNL, and Sandia National Laboratories designed to support collaborative projects with external partners in government, industry, and academia. The Lab’s predictive biology research includes drug design modeling as well as machine learning for COVID-19 antibody designs.

Industry Forum Recap
LLNL’s first-ever Machine Learning for Industry Forum (ML4I) was held on August 10–12. The virtual event was sponsored by the HPCIC and the DSI. The Forum’s goals were to encourage and illustrate the adoption of ML methods for practical outcomes in various industries, particularly manufacturing. Discussions, panel sessions, and presentations were organized around three high-level topics: industrial applications, tools and techniques, and ML’s impact and potential in industry.
Acting HPCIC director Wayne Miller explained, “There is a need to develop collaborations between our data scientists who ‘know how to make ML work’ and industry users who have data but not much experience in developing ML tools.” DSI director Michael Goldman added, “The DSI’s research and outreach efforts complement the HPCIC’s computational resources and expertise. It made sense to partner with each other on this Forum.”
Highlights included keynote speakers from industry, academia, and government; talks on ML computing workflows, RL for simulations, and cognitive simulation; and panel discussions on the ML workforce and dataset sharing and security. About 50 presenters answered the call for abstracts in April, and the event’s large turnout and audience response make an ML4I Forum likely next year. A recap of the event is available on the DSI website, and presentations can be downloaded here.

Virtual Seminar Series Continues into FY22
LLNL’s new fiscal year begins in October and marks the beginning of the DSI’s fourth year of hosting its seminar series. Since going virtual in 2020, the DSI has invited 16 speakers to share their research on topics ranging from deep symbolic regression and stochastic simulation experiments to low-dimensional modeling and natural language processing. Behind the scenes, the series is coordinated by a member of the Lab’s technical staff as well as DSI administrator Jen Bellig. For FY22, statistician and series coordinator Kathleen Schmidt will pass the baton to Sarah Mackay, a postdoctoral researcher at LLNL’s Center for Applied Scientific Computing. Mackay joined the Lab in 2019 and holds a PhD in Mathematics from the University of Illinois at Urbana-Champaign. (Image at left: LLNL’s Brenden Petersen delivered a virtual seminar titled “Deep Symbolic Regression: Recovering Mathematical Expressions from Data via Risk-Seeking Policy Gradients.”)

Virtual Seminar: Gaussian Process Models
For the DSI’s August 26 virtual seminar, Dr. Amanda Muyskens presented “MuyGPs: Scalable Gaussian Process Hyperparameter Estimation Using Local Cross-Validation.” She introduced attendees to MuyGPs, a novel computationally efficient Gaussian process hyperparameter estimation method for large data. The method has been applied to space-based image classification and released for open-source use in the Python package MuyGPyS. Muyskens works in LLNL’s Applied Statistics Group and holds a PhD in Statistics from NC State University. A recording of her talk will be posted to the seminar series’ YouTube playlist.
R&D Highlights
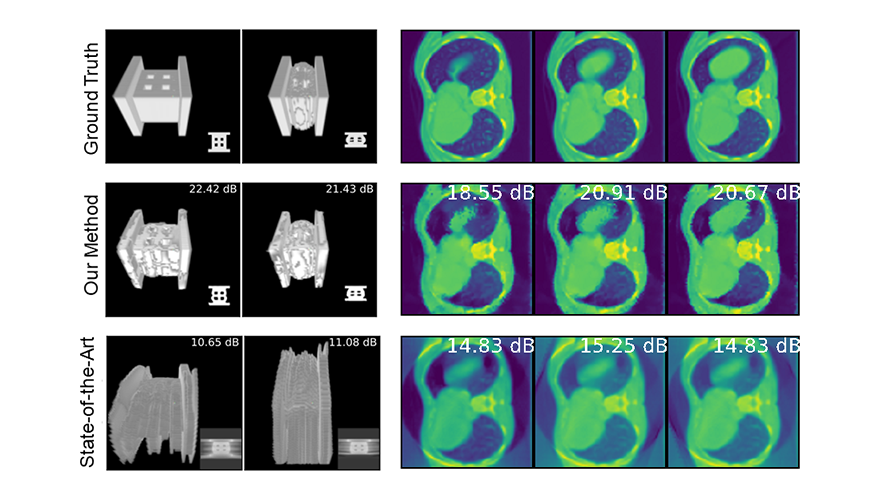
New derivation for linear dynamical systems. A classical reduced order model (ROM) for dynamical problems typically involves only the spatial reduction of a given problem. In a paper recently published in Mathematics, LLNL researchers and a University of California, Berkeley, colleague present for the first time the derivation of the space–time least-squares Petrov–Galerkin projection for linear dynamical systems and its corresponding block structures. (Image at left: From a recent Mathematics paper, examples of a 2D linear diffusion equation solving speed-ups versus reduced dimensions for Galerkin projection [left] and least-squares Petrov–Galerkin projection [right].)
ML innovation in cancer research. LLNL’s relationship with cancer research endures some 60 years after it began, with historical precedent underpinning exciting new research areas. The latest cover story of Science & Technology Review describes a range of research including interactions between the RAS protein and a cell’s lipid membrane. Using high-performance computers, scientists have run tens of thousands of simulations to better understand RAS interactions and activities with proteins and the lipid membrane. The massively parallel Multiscale Machine-Learned Modeling Infrastructure (MuMMI) has helped scientists find relevant seed data for further modeling and analysis of RAS protein interactions.
Scalable visualization software. The recent issue of Science & Technology Review also includes an article about VisIt, which has become a trusted tool since its development began in 2000. VisIt is open-source software that enables researchers to rapidly visualize, animate, and analyze scientific simulations. It is the Lab’s go-to software for scalable visualization as well as graphical and numerical analysis of both simulated and experimental datasets.

Meet an LLNL Data Scientist
Katie Lewis joined LLNL in 1998 after earning a B.S. in Mathematics from the University of San Francisco. She has held numerous leadership positions in the Computing, National Ignition Facility, and Weapons and Complex Integration directorates. Today, Lewis serves as Associate Division Leader for the Computational Physics Section of the Design Physics Division and leads the Vidya ML project in LLNL’s Weapons Simulation and Computing Program, where she applies AI techniques to HPC simulations. “AI/ML is proving to be a gamechanger in approaching challenging problems related to scientific computing. Employing these techniques to solve problems more accurately and more quickly will lead to greater scientific discovery,” she states.
Learn About Reduced Order Models
ROMs combine data and underlying first principles to accelerate physical simulations, reducing computational complexity without losing accuracy. An LLNL team has developed an open-source C++ software library called libROM, which provides data-driven physical simulation methods from intrusive projection-based ROMs to non-intrusive black-box approaches. Computational scientist Youngsoo Choi recorded two tutorials for libROM users: