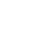June 15, 2020
Spotlight: Materials Science Meets AI
LLNL scientists have taken a step forward in the design of future materials with improved performance by analyzing its microstructure using AI. The work recently appeared in the journal Computational Materials Science.
Technological progress in materials science applications spanning electronic, biomedical, alternate energy, electrolyte, catalyst design, and beyond is often hindered by a lack of understanding of complex relationships between the underlying material microstructure and device performance. But AI-driven data analytics provide opportunities that can accelerate materials design and optimization by elucidating processing-performance correlations in a mathematically tractable way.
Recent developments in artificial-neural-network-based deep learning methods have revolutionized the process of discovering such intricate relationships using the raw data itself. However, to reliably train large networks one needs data from tens of thousands of samples, which, unfortunately is often prohibitive in new systems and new applications due to the cost of sample-preparation and data collection.
Innovative algorithms are needed to extract the most appropriate “features” or “descriptors” out of the raw experimental characterization data. A team of materials scientists and data-visualization scientists at LLNL and the University of Utah used recently developed methods in scalar-field topology and Morse theory to extract useful summary features like “grain count” and “internal boundary surface area” from the raw X-ray computed tomography data. (Image: X-ray CT images of materials created from five different lots.)
Spotlight: Introducing the COVID-19 Data Portal
To help accelerate discovery of therapeutic antibodies or antiviral drugs for SARS-CoV-2, the virus that causes COVID-19, LLNL has launched a searchable data portal to share its COVID-19 research with scientists worldwide and the general public.
The portal houses a wealth of data LLNL scientists have gathered from their ongoing COVID-19 molecular design projects, particularly the computer-based “virtual” screening of small molecules and designed antibodies for interactions with the SARS-CoV-2 virus for drug design purposes. The data is queryable by criteria such as chemical structure and binding probability scores, so outside researchers can easily locate relevant data for their own work.
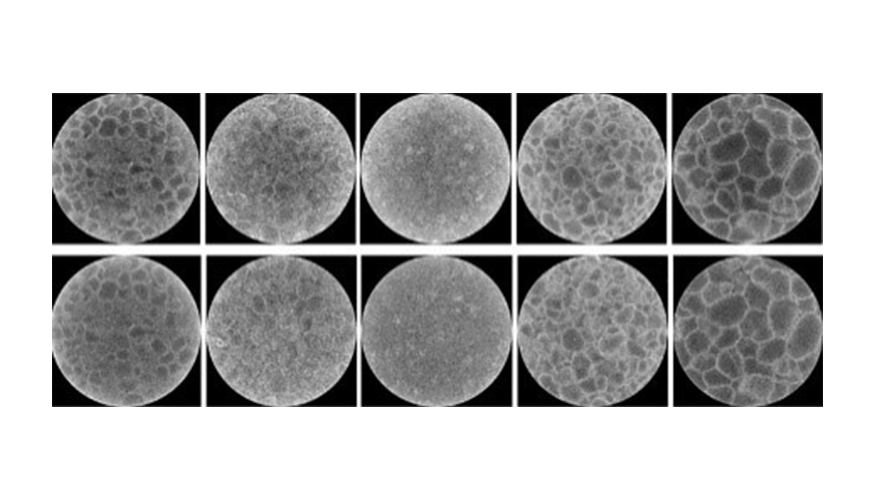
The portal will be regularly updated and will, in a few months, provide the results of experiments performed at the Laboratory on the effectiveness of small molecules and antibodies against SARS-CoV-2. (Image: Protein protease structure.)
Recent Research
- In additional materials science research, a team has developed machine learning tools that extract and structure information from the text and figures of nanomaterials articles using state-of-the-art natural language processing, image analysis, computer vision, and visualization techniques.
- LLNL scientists continue to contribute to machine learning research by expanding on calibration techniques. A new paper—recently accepted to the upcoming 37th International Conference on Machine Learning—studies the problem of post-hoc calibration of ML classifiers. The authors demonstrate "Mix-n-Match" calibration strategies (i.e., ensemble and composition) that help achieve remarkably better data efficiency and expressive power.
- A group of LLNL data scientists have helped arrange an applied machine learning track for the August 2020 SPIE Optical Engineering and Applications Conference in San Diego. Conference chair is LLNL’s Michael Zelinski.
The Fight Against COVID-19
- A team led by Jay Thiagarajan has come up with a new approach for improving the reliability of artificial intelligence and deep learning-based models used for critical applications, such as health care. They recently applied the method to study chest X-ray images of patients diagnosed with COVID-19.
- Researchers have identified an initial set of therapeutic antibody sequences, designed in a few weeks using machine learning and supercomputing, aimed at binding and neutralizing SARS-CoV-2, the virus that causes COVID-19. The research team is performing experimental testing on the chosen antibody designs.
- LLNL’s coincidentally named Corona supercomputer has been upgraded for COVID research, with new processors designed for deep learning.

Multimedia Highlights
- LLNL director Bill Goldstein was featured on the Hidden in Plain Sight podcast in the episode “Using Data to Build a Secure Future,” discussing the importance of data analysis to the Lab’s mission.
- Jay Thiagarajan was featured on the Data Skeptic podcast in an episode called “Calibrating Healthcare AI.” He described the challenges of interpreting machine learning models.
- At the Stanford HPC Conference, Katie Lewis talked about incorporation of machine learning—one of the fastest growing areas of computing—into scientific simulations at LLNL.
- In a Faces of STEM video, Brenda Ng explained why she loves her job and what inspired her to pursue a career in STEM.
Workforce Updates
The DSSI’s annual program will be conducted online this year. Twenty-six students will begin their 12-week internships in June.
For the second year in a row, the DSI has teamed up with the University of California at Merced to offer a two-week Data Science Challenge at the beginning of the summer. The intensive program provides mentors, assignments, virtual tours, and seminars. Under the direction of LLNL’s Marisol Gamboa and UC Merced’s Suzanne Sindi, 21 students are applying data science techniques to a materials science project.
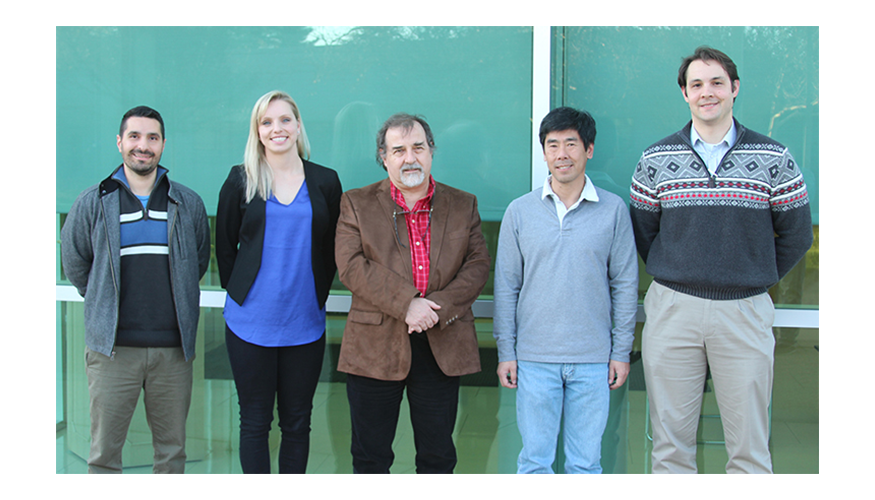
Meet a Research Team
LLNL researchers are developing 3D protein structures that predict the new virus’s protein structure, which is currently unknown. The models combine the COVID-19 genomic sequence with the known structure of a protein found in the virus that causes severe acute respiratory syndrome SARS—another type of coronavirus. The LLNL team is, left to right, Daniel Faissol, Magdalena Franco, Adam Zemla, Edmond Lau, and Thomas Desautels.